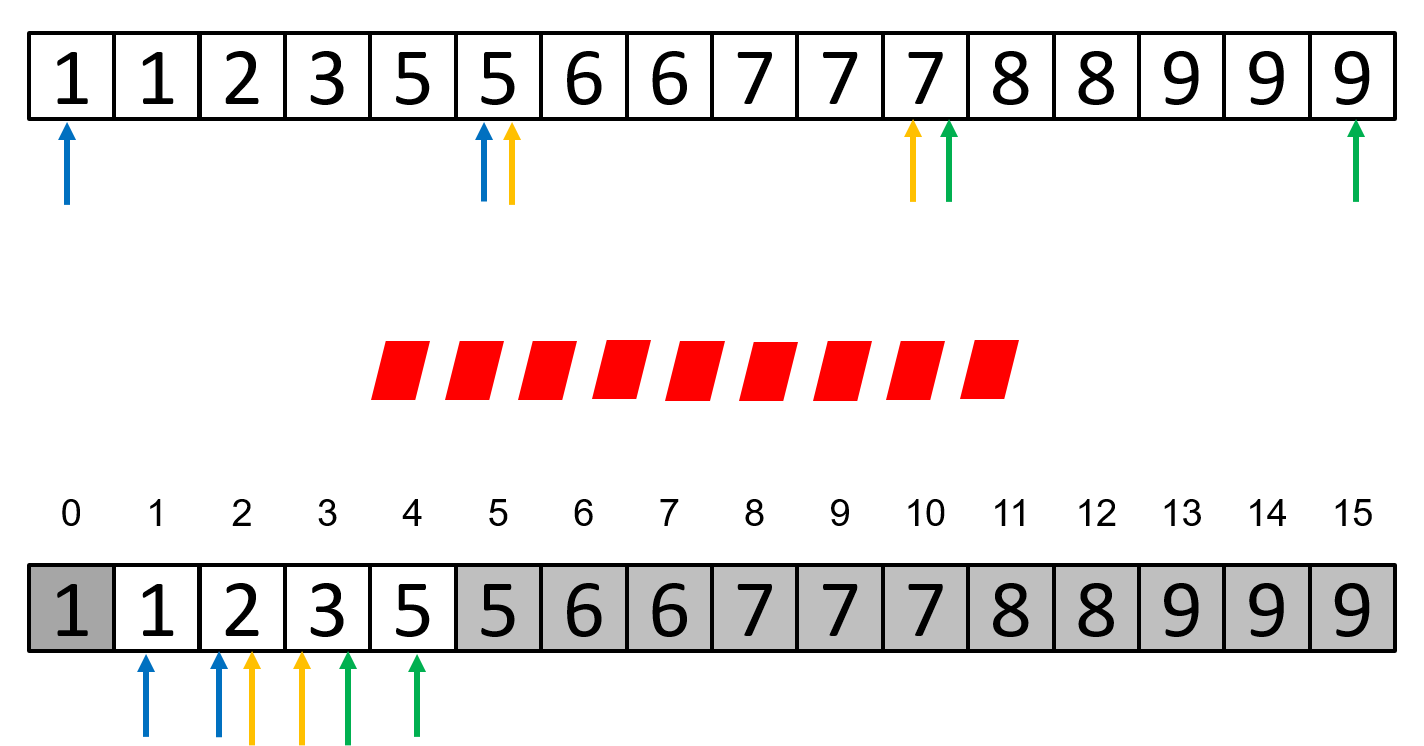
Parallel Binary Search
The parallel binary search algorithm is based on dividing the sequence in which the search is performed into P subsequences of the same length. At each step, each of the P processors checks whether a value x is present in the subsequence assigned to that processor by comparing the value of x with the values at the start and end positions, marking the boundaries of the processor's subsequence.
If a processor finds the value x in its subsequence, it will notify the other processors about this, and then all P processors will search in that subsequence, which will be divided into other P subsequences, and the search will proceed to the next step.
Unlike the serial version, where it checks if the searched element is in the middle position of the sequence, in the parallelized version, it checks if the element is in one of the start and end positions of the sequence (if it is in one of these positions, it means that we have found the element in the sequence).
It can be observed that the operations of searching for the value x in subsequences by each processor at each step can be performed in parallel. However, all search operations in one step must be completed entirely before moving to the next step, so in this case, a barrier is required after each search step.
A graphical representation of the parallel binary search algorithm can be seen in the image below, where operations with the same color can be performed in parallel, lines of the same color represent the start and end indices of each subsequence, specific to each processor, and the symbols in red represent barriers.